
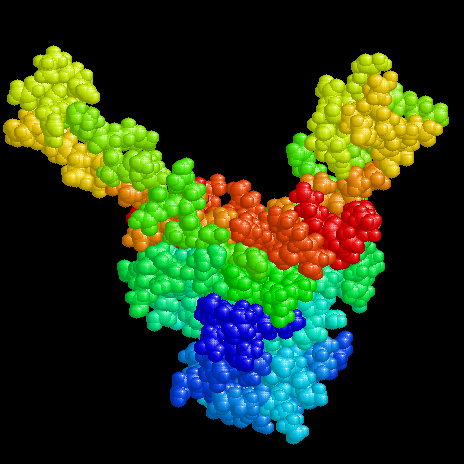
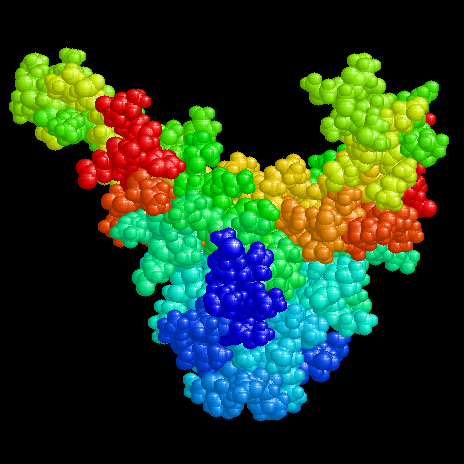
The linear textbook structure of DNA is neither apt for storage in a living cell nor for function during transcription or replication. Within a cell DNA has to be folded, wich is accomplished in eukaryotes e.g. by nucleosomes. Also the chromosome of Escherichia coli with a length of ca. 1 mm has to be brought to a compact form to fit in a space of about 1 Ám diameter. In bacteria there were found a couple of histone like proteins fulfilling this task as well as those needed in transcription and replication. The molecular structure of some of these proteins and their complexes with DNA have been investigated in the crystalline state by x-ray or in solution by NMR.
The proteins shown here are about 20 kDa in size and exist as homologous hetero dimers (IHF) or homo dimers (HU, TF1). They bind nonspecifically to DNA (HU) or to defined consensus sequences (IHF, TF1). Their structures are characterized by compact 'bodys' from which 'arms' emerge, which interact with the DNA.
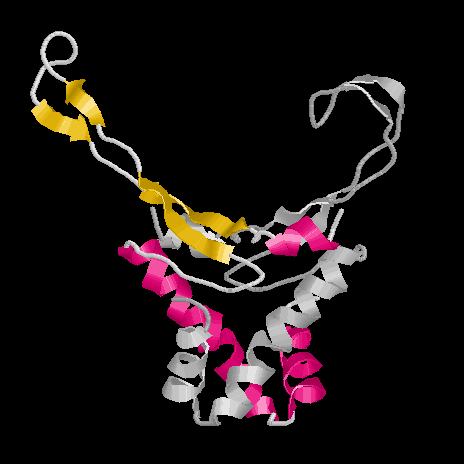 In this figure one subunit of HU is depicted in grey, the other is colored according to its structure: alpha-helices forming the 'body' are red and beta-sheets within the 'arms' are shown in yellow. The arms each contain two beta-sheets with a flexible region in between. This allows a folding of the arms into the minor groove of the DNA.
In this figure one subunit of HU is depicted in grey, the other is colored according to its structure: alpha-helices forming the 'body' are red and beta-sheets within the 'arms' are shown in yellow. The arms each contain two beta-sheets with a flexible region in between. This allows a folding of the arms into the minor groove of the DNA.
This structure was elucidated by NMR from dissolved HU without bound DNA.
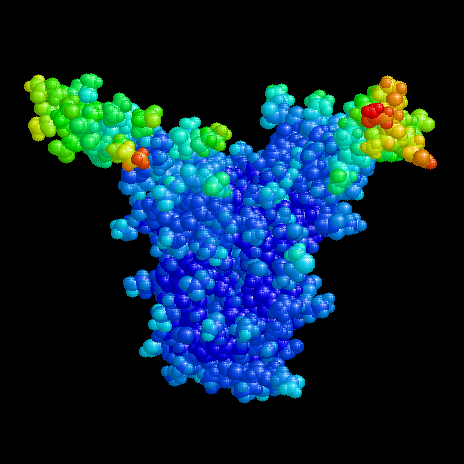 Here TF1 is shown to demonstrate the flexibility of the arms. The data originate from x-ray diffraction within a protein crystal. In the diffraction pattern each atom is positioned with a precision resulting from relative deviations in individual molecules. This blurriness is described for every atom in the protein data bank as the temperature factor. So there are 'cold' atoms (rigidly bound) and 'hot' atoms with some degree of movability even in the crystalline state. The figure shows cold atoms in blue and hot atoms in red. The tips of the arms have the highest degree of flexibility according to the temperature factor.
Here TF1 is shown to demonstrate the flexibility of the arms. The data originate from x-ray diffraction within a protein crystal. In the diffraction pattern each atom is positioned with a precision resulting from relative deviations in individual molecules. This blurriness is described for every atom in the protein data bank as the temperature factor. So there are 'cold' atoms (rigidly bound) and 'hot' atoms with some degree of movability even in the crystalline state. The figure shows cold atoms in blue and hot atoms in red. The tips of the arms have the highest degree of flexibility according to the temperature factor.
At first IHF (integration host factor) was recognized to be neccessary for the integration of phage lambda into the chromosome of E. coli. Later IHF was found to function both as a positive or negative regulator in transcription in different parts of the genome (Review: N Goosen, P vd Putte, Mol. Microbiol. 16 (1996) 1-7). In each case the function of IHF is to sterically prepare the DNA for other proteins to interact in specific spots.
IHF was crystallized together with a consensus DNA fragment. Its interaction with DNA is a good example for this class of proteins.