The following lists all nucleosides found in tRNAPhe
 |
|
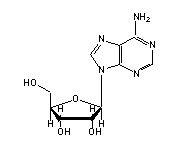
Adenosine (A, 5,9,13,14,21,23,29,31,35,36,38,39,44,62,64,66,67,73,76) in tRNA: | |
 |
|
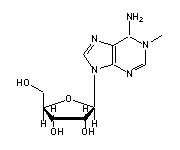
1-methyladenosine (1MA, 58) in tRNA: | |
 |
|
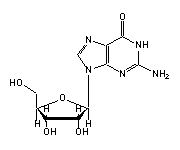
Guanosine (G, 1,3,4,15,18-20,22,24,30,42,43,45,51,53,57,65,71) in tRNA: | |
 |
|
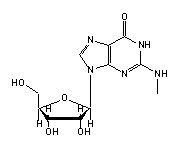
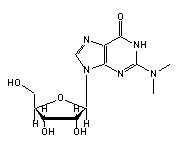
N2-methylguanosine (2MG, 10) in tRNA: | |
 |
|
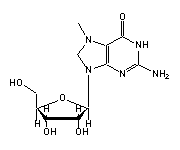
N2-dimethylguanosine (M2G, 26) in tRNA: | |
 |
|
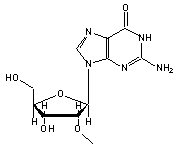
7-methylguanosine (7MG, 46) in tRNA: | |
 |
|
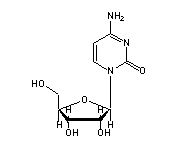
O2'-methylguanosine (OMG, 34) in tRNA: | |
 |
|
Cytidine (C, 2,11,13,25,27,28,48,56,60,61,63,70,72,74,75) in tRNA: | |
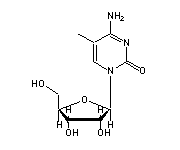 |
|
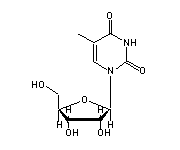
5-methylcytidine (5MC, 40,49) in tRNA: | |
 |
|
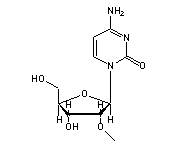
O2'-methylcytidine (OMC, 32) in tRNA: | |
 |
|
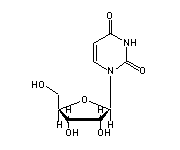
Uridine (U, 6-8,12,33,41,49,50,52,59,68,69) in tRNA: | |
 |
|
5-methyluridine (5MU, 54) in tRNA: | |
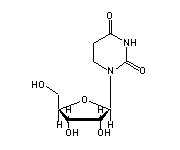 |
|
5,6-dihydrouridine (H2U, 16,17) in tRNA: | |
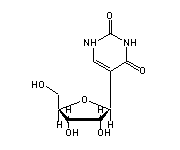 |
|
Pseudouridine (PSU, 39,55) in tRNA: | |
 |
|
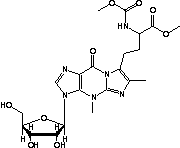
Wybutosine (YG, 37) in tRNA: | |
Basepairings in tRNAPhe are:
Unpaired bases
The 'arm' designation originates from the time when only the secondary structure was known ('cloverleaf') and is somewhat misleading in the tertiary structure
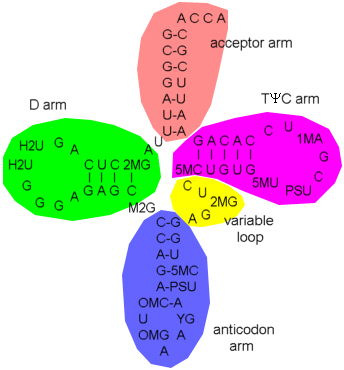
The structures marked red and dark green in the left frame
The backbone of the tRNA is partly stabilized by water molecules as shown here for the T psi C arm (G51 - U59)
The most rigid part of the tRNA is the central domain formed by the D arm and the T psi C arm (which are folded tightly together)
Some twin water molecules bridge the ribose 2' hydroxyl group to a phosphate oxygen of the next nucleotide
H Shi & PB Moore, The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited, RNA 6 (2000) 1091-1105
L Jovine et al, The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: Cleavage by Mg2+ in 15-year old crystals, J. Mol. Biol. 301 (2000) 401-414
10-01 - R Bergmann