| From Sequence to FunctionAn Introduction to the KEGG Project |
|---|
The functional data that relate to sequence information are currently stored, for example, in the features tables of the sequence databases and in the motif libraries such as PROSITE. However, these basically represent sequence-function relationships of single molecules, i.e., individual components of a biological system, and they do not contain higher level information, i.e., wiring diagrams, of genetic interactions and molecular interactions.
We have thus initiated the project named KEGG (Kyoto Encyclopedia of Genes and Genomes), first, to computerize current knowledge of molecular and cellular biology in terms of the information pathways that consist of interacting genes or molecules and, second, to link individual components of the pathways with the gene catalogs being produced by the genome projects.(*1) Despite its naming of encyclopedia, KEGG is not simply a static resource to be searched and browsed. KEGG is a deductive database in the sense that additional information can be deduced dynamically from the stored information. Although such deductive capabilities have not been fully implemented yet, we hope the current version of KEGG will still be able to assist logical reasoning processes of functional assignments from sequence data.
Pathway maps are the main feature of KEGG, which is the collection of graphical diagrams representing the information pathways of interacting molecules or genes. KEGG contains all known metabolic pathways and a limited, but increasing, number of regulatory pathways.
Molecular catalogs are intended to represent functional aspects of proteins, RNAs, other biological macromolecules, small chemical compounds, and their assemblies. The current version of KEGG contains four tables of enzyme classifications.
Gene catalogs contain classifications of all known genes for each organism. Depending on how one views the function, genes may be classified in a number of different ways. KEGG provides the classification scheme according to the pathway information, as well as other schemes by different authors.
Genome maps are presented to help understand the physical locations of genes and their relationship with the pathways, as well as to assist handling of genes. Genome maps are manipulated graphically by Java.
DBGET is the backbone of the GenomeNet database service and currently supports the following databases:
The PATHWAY and GENES databases are the results of the KEGG project. In addition, the construction and maintenance of LIGAND is now undertaken as a part of the KEGG project, in collaboration with Takaaki Nishioka who originally developed the database (Suyama et al., CABIOS 9, 9-15, 1993).
In KEGG each diagram has been drawn and is continuously updated manually.
A diagram for the metabolic pathways does not represent a consensus of known pathways in different
organisms; it is intended as a reference drawing of all chemically feasible pathways.
The organism-specific pathways are then automatically generated by
matching the enzyme genes in the gene catalog with the enzymes on the reference
pathway diagrams.
In contrast, a diagram for the regulatory pathways is drawn
separately for each organism.
The collection of pathway diagrams forms the PATHWAY database in KEGG
that can also be handled in the DBGET/LinkDB system.
The standard (reference) pathway diagrams in the PATHWAY database are linked
to the LIGAND database that consists of the ENZYME section for enzyme reactions
and the COMPOUND section for metabolic compounds.
All reactions and compounds in the PATHWAY database should be in the
LIGAND database, but this is not complete yet.
The organism specific pathway diagrams are linked to the GENES database,
which is a collection of gene catalogs for individual organisms
containing sequence and other information.
The functional hierarchy of genes in the GENES database can be handled
by the KEGG hierarchical text representation.
There are two versions of the hierarchy, the KEGG version based on
the pathway classification and the original version provided by the authors
of the genome sequencing.
The latter is often based on Monica Riley's classification scheme.
The KEGG pathways can also be searched by sequence similarity.
This is especially useful for identifying orthologues and reconstructing
pathways from the gene catalog.
For example, by taking the E. coli pathways as references, the user
can check if a functional unit can be formed from the gene catalog of
a specific organism.
Perhaps, the most challenging task in KEGG is the inference capabilities
that will help human to make logical reasoning.
Given a list of enzymes (EC numbers) that are found in the gene catalog of
an organism, KEGG automatically generates the organism specific pathways
by marking the matching enzymes on the diagram.
Then, the connectivity and completeness of the marked enzymes can be used
to assess the correctness of functional assignment in the gene catalog.
The existence of a missing element implies either the gene catalog is wrong
or there is an unknown reaction pathway that utilizes different enzymes
in the catalog.
For the latter possibility, KEGG provides an option to compute pathways
from a given list of enzymes. This is done by deduction from binary
relations of substrates and products with an optional use of query relaxation
for functional hierarchies.
For the former possibility, it is necessary to develop a gene finding
and functional prediction system that incorporates the knowledge of
reconstructed pathways. The current version of KEGG provides an experimental
server for automatic assignment of EC numbers from a list of all protein sequences
in the whole genome based on the relations of orthologous genes.
We are currently working on to improve the performance of the CD version
by limiting certain capabilities that are available in the Internet version.
Please direct your comments to www@genome.ad.jp.
Last Updated: December 11, 1997
(*1) KEGG is a chicken (pathway) or egg (gene catalog) problem.
Data Representation
From the user's point of view, the data in KEGG are represented either by
graphical diagrams (pathway maps and genome maps) or hierarchical texts
(gene catalogs and molecular catalogs).
Internally, however, the most basic data item is the binary relation;
other data types are considered the composites of them (see figure below).
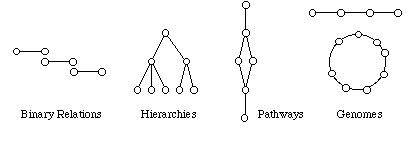
The binary relations are especially useful in comparing and computing
pathways, genomes, and hierarchies.
For example, KEGG provides the capability of computing all possible
metabolic pathways from a given list of binary relations between
substrates and products, eg., a list of enzymes.
The concept of binary relation has also been successfully implemented in the
LinkDB database of the DBGET system,
where the user can deduce additional relations by computing on multiple relations.
Data Collection
KEGG currently contains most of the known metabolic pathways and some of the
known regulatory pathways represented by about 100 graphical diagrams.
The metabolic pathways were originally compiled from the book "Metabolic Maps" by the Japanese
Biochemical Society and the Boehringer wall chart of "Biochemical Pathways".
We have started collaborating and cross-linking with
WIT which provides a more detailed
picture of the metabolic pathways.
In contrast, KEGG attempts to cover a wider range of biochemical pathways
at a higher level of abstraction.
Starting in July 1997 the regulatory pathways are also made available in KEGG.
They are mostly compiled from the primary literature.
We welcome any form of collaboration to organize or verify these pathways.
Search and Compute
The KEGG pathways can be searched by EC numbers for enzymes, by compound
numbers for chemical compounds, and gene accessions for specific genes.
If the search is combined with the KEGG grouping or the hierarchical
classification, this is as if performing a relational join operation.
For example, by taking the EC numbers from a specific group in the
superfamily table (or the SCOP table) and searching them against the
pathway diagrams, the user can view immediately whether there is
a tendency of similar genes appearing in cluster on the pathway, i.e.,
an indication of gene duplications in the pathway formation.
Technical Notes
KEGG is released in two versions, the Internet version and the
CD version,
both of which are to be browsed by an Web browser, such as Netscape Navigator
and Microsoft Internet Explorer.
The programs to handle the four data types in KEGG are written either by the
CGI (Common Gateway Interface) scripts or by Java.
As can be seen in the table above, the browser must be Java compatible
to use the CD version, but the Internet version can be used without Java
as long as the genome maps are not necessary for the user.
Data type \ Version Internet CD
Pathway map CGI Java
Molecule table CGI Java
Gene table CGI Java
Genome map Java Java
Created: November 28, 1995
