The bc1-complex
Complex III
|
Name
Formal name: ubiquinol:cytochrome c oxidoreductase
Trivial name: bc1-complex (bc-complex)
Alternative name: Complex III
Function
The formal name describes the function: the enzyme oxidizes ubiquinol (ubihydroquinone)
which reacts from the membrane phase, reduces cytochrome c in the intermembrane
space (or periplasm in bacteria), and uses the free energy change to transport
2 (vectorial) H+/QH2 across the membrane from matrix
(N side) to inter membrane space (P side), and release 2 additional (scalar)
H+/QH2 into the inter membrane space.
QH2 + 2 ferricyt c3+ + 2H+N
<==> Q + 2 ferrocyt c2+ + 4H+P
Mechanism
The bc1-complex works through a modified
Q-cycle mechanism. The link has details of the reaction mechanism as
determined for the enzyme in the photosynthetic chain of Rb. sphaeroides.
In addition, physico-chemical data for redox parameters, rate constants,
etc., which describe the mechanism are given.
The Q-cycle mechanism is summarized in the following Scheme:
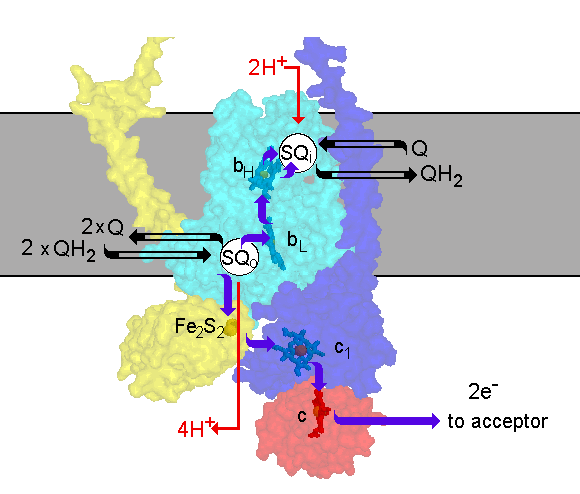 The scheme shows the Q-cycle in the context of the structure. The catalytic
subunits are shown by their surfaces, made transparent so as th reveal
the redox centers. The reactions are catalyzed by three subunits; cyt b
(cyan), containing two b-type hemes, bL (lower potential heme)
and bH (higher potential heme); cyt c1 (blue), containing
heme c1; and the iron sulfur protein (yellow), containing an
Fe2S2 center. Quinol is oxidized at the Qo-site
of the complex. The reaction is rate determining and has a relatively high
activation barrier. Quinol oxidation occurs in a bifurcated reaction, in
which one electron is transferred to a high potential chain and the other
to a low potential chain. The high potential chain, consisting of the ISP,
cyt c1 and cyt c (or c2) (red), transfers the first
electron from quinol to an acceptor (cytochrome oxidase in mitochondria,
the oxidized photochemical reaction center in photosynthetic systems),
leaving a semiquinone at the Qo-site. Because the semiquinone
formed is unstable, and undetectable during normal turnover, the reaction
at the Qo-site appears to be a concerted electron transfer to
the high and low potential chains. The low potential chain consists of
two cyt b hemes (cyt bL and cyt bH, for low and higher
potential hemes), which serve as a pathway through which electrons are
transferred across the coupling membrane from semiquinone at the Qo-site
to the Qi-site, at which quinone is reduced to quinol. In order
to provide the two electrons at the Qi-site required for reduction
of quinone, the Qo-site oxidizes two equivalents of quinol in
successive turnovers. The first electron at the Qi-site generates
a relatively stable semiquinone which is reduced to quinol by the second
electron. The integration of the oxidation and reduction reactions with
the release or uptake of protons in the aqueous phases, allows the complex
to pump protons across the membrane. Electron transfer between the two
Q-sites through the b-cytochrome chain provides the electrogenic work.
The scheme shows the Q-cycle in the context of the structure. The catalytic
subunits are shown by their surfaces, made transparent so as th reveal
the redox centers. The reactions are catalyzed by three subunits; cyt b
(cyan), containing two b-type hemes, bL (lower potential heme)
and bH (higher potential heme); cyt c1 (blue), containing
heme c1; and the iron sulfur protein (yellow), containing an
Fe2S2 center. Quinol is oxidized at the Qo-site
of the complex. The reaction is rate determining and has a relatively high
activation barrier. Quinol oxidation occurs in a bifurcated reaction, in
which one electron is transferred to a high potential chain and the other
to a low potential chain. The high potential chain, consisting of the ISP,
cyt c1 and cyt c (or c2) (red), transfers the first
electron from quinol to an acceptor (cytochrome oxidase in mitochondria,
the oxidized photochemical reaction center in photosynthetic systems),
leaving a semiquinone at the Qo-site. Because the semiquinone
formed is unstable, and undetectable during normal turnover, the reaction
at the Qo-site appears to be a concerted electron transfer to
the high and low potential chains. The low potential chain consists of
two cyt b hemes (cyt bL and cyt bH, for low and higher
potential hemes), which serve as a pathway through which electrons are
transferred across the coupling membrane from semiquinone at the Qo-site
to the Qi-site, at which quinone is reduced to quinol. In order
to provide the two electrons at the Qi-site required for reduction
of quinone, the Qo-site oxidizes two equivalents of quinol in
successive turnovers. The first electron at the Qi-site generates
a relatively stable semiquinone which is reduced to quinol by the second
electron. The integration of the oxidation and reduction reactions with
the release or uptake of protons in the aqueous phases, allows the complex
to pump protons across the membrane. Electron transfer between the two
Q-sites through the b-cytochrome chain provides the electrogenic work.
You can download a working
model of the bacterial photosynthetic electron transfer chain, showing
the Q-cycle, or view the model
in action (be patient when linking to this page, because the full model
will take a while to load). A more recent model based on suggestions from
the Crofts Lab for a molecular mechanism, and the recent structural information
from Ed Berry's lab, is shown here in a 3-D
movie.
Subunit composition
The bc1-complex from beef heart mitochondria
Subunit (no.) Redox centers Mr(beef) Function
Core I none 53.6 No catalytic, protein transport
Core II none 46.5 No catalytic, protein transport
Cyt b (III) heme bH 42.6 donor to Qi-site
heme bL acceptor from SQ at Qo-site
Transmembrane electron transfer
Cyt c1(IV) heme c1 27.3 donor to cyt c
Rieske (V) 2Fe.2S center 21.6 acceptor from QoH2
donor to cyt c1
Subunit VI none 13.3 none known
Subunit VII none 9.5 none known
Subunit VIII none 9.2 hinge protein (interacts with c1)
Subunit IX none 8.0 none known
Subunit X none 7.2 none known
Subunit XI none 6.4 none known (not present in chicken)
The bc1-complex from Rhodobacter sphaeroides
Cyt b (I) heme bH 50 donor to Qi-site
Acceptor of electrons from heme bL
heme bL acceptor from SQ at Qo-site
Transmembrane electron transfer
Cyt c1(II) heme c1 28.6 donor to cyt c
Rieske (III) 2Fe.2S center 19.9 acceptor from QoH2
donor to cyt c1
Subunit IV none 14.4 may contribute to Qo-site
Sequence information
Sequences for all subunits
of the beef heart mitochondrial complex have been compiled by Ed Berry,-
other sequences are accessible from Ed's
home page.
Click on these links for alignments of sequences from many
mitochondrial, and some
bacterial bc1-complexes.
The structure of the bc1-complex
Structures of the bc1 complex from mitochondria have been published
by three groups, the first from a collaboration between Diesenhofer's group
and Chang-An Yu's group (Xia et al., 1997) (1), and from Berry's group
in Kim's laboratory (Zhang et al., 1998) (2). More recently, anothe structure
of the beef complex has been published by Iawata et al. (10). All complexes
so far solved are dimeric, showing a two-fold symetry about an axis vertical
to the membrane plane; all groups report crystals with a homodimer containing
two bc1-complex monomers with a subunit composition as described
in the table above. There is enough interdigitation between monomers to
suggest that dissociation of the dimer would be unlikely. It therefore
seems probable that the complex is structurally dimeric in its native state.
The structure of mitochondrial complexes from several sources have been
solved in two laboratories (see refs. below). The structure of the complete
chicken heart mitochondrial complex from Ed Berry's work (Zhang et al.,
1998) (2) is shown here:
 Click thumbnail for larger version
Click thumbnail for larger version
General features
The general features of the protein are as expected from biochemical and
previous structural studies, with a large fraction corresponding to the
"core" proteins (subunits I and II) on the N-side (bottom in the Fig. above),
and cyt c1 and the FeS protein on the P-side. The dimensions
of the dimer are about 130 Å in diameter and 151 Å in height,
with the inter-membrane space region, the transmembrane region, and the
matrix region contributing about 41 Å, 35 Å and 75 Å
respectively.
Prosthetic groups
The prosthetic group composition is as expected from biochemical studies:
cyt bL, cyt bH, cyt c1, 2Fe.2S center.
Cyt bL --> cyt bH distance is 20 Å (13
Å edge to edge), perpendicular to membrane. Assuming that cyt bH
is close to the antimycin binding site, the arrangement in the protein
is as expected from models based on mechanistic studies and structural
prediction.
The positions of the heme centers are essentially the same in all structures.
The cyt bL hemes of the two monomers are 21 Å apart in
the dimer, the cyt bH hemes are 33 Å apart, and the cyt
c1 hemes are 53 Å apart.
In the Xia et al. structure (1, not shown), the protein of the iron
sulfur protein (ISP) was not resolved, because of disorder; the cyt c1
subunit was only partly resolved. The position of the 2Fe.2S center was
determined, with the following distances:
Cyt L --> 2Fe.2S distance is 26 Å.
2Fe.2S --> cyt c1 distance is 31 Å
the FeS centers are 63 Å apart.
In the Zhang et al. (2) structure, both the ISP and cyt c1
were well resolved, but showed different distances between the Fe-centers
in the native crystals:
Cyt L --> 2Fe.2S distance is 34.3 Å.
2Fe.2S --> cyt c1 distance is 21.3 Å
When the structure of crystals containing the Qo-site inhibitor
stigmatellin were solved (2), the position of the ISP had changed so as
to be close to that inferred from the Xia et al. (1) distances. Distances
between the Fe-centers in the stigmatellin crystals were:
Cyt L --> 2Fe.2S distance is 26.4 Å.
2Fe.2S --> cyt c1 distance is 31.6 Å
Zhang et al. (1) and Crofts et al. (3) have suggested that the two different
positions reflect a domain movement which is essential for catalysis, since
in neither of the positions observed would the complex be fitted for catalysis
of all the partial reactions of quinol oxidation.
Spatial considerations
The biggests surprise from the earlier Xia et al. (1) structure was the
distance between the 2Fe.2S center and cyt c1. The kinetic data
suggest that this reaction is rapid, with t½ < 10
µs. The distance of 31 Å observed in the Xia et al. structures
would not
permit this rate. The distances between centers in Berry's native structures
(2) are similar to those in the Xia/Deisenhofer/Yu structure, except for
the position of the 2Fe.2S centers. These are closer to cyt c1,
and further from heme bL. These differences have led is to suggest
that the extrinsic domain of the Rieske ISP containing the 2Fe.2S center
(the mobile head) must move relative to the other two catalytic subunits,
by a rotational displacement of ~22 Å. The movement of the ISP head
brings the 2Fe.2S cluster from a position close to cyt c1 to
a concave interface on cyt b at which it is close to the quinol binding
site (the Qo-site). In the presence of stigmatellin, the contact
seen in electron density maps can be modeled as a H-bond between Ne
of His-161 (a ligand to the 2Fe.2S center), and a carbonyl- and a methoxy-
O-atom of the stigmatellin ring (2).
There are 13 transmembrane helices apparent in the X-ray structure.
Eight of these belong to cyt b (SUIII), as in models based on structural
prediction. The remaining four helices are assigned to the remaining subunits,
one each coming from cyt c1 (SUIV), ISP (SUV), SUVII and SUX)
(1, 2).
In addition to the transmembrane helices, cytochrome b has five prominent
helices outside the membrane, and parallel to the membrane plane. These
correspond to amphipathic helices a, ab, cd, and ef predicted from sequence
analysis, with the cd span forming two helices, cd1 and cd2 in a hairpin,
with a turn at the conserved proline. The arrangement of transmembrane
helices around the hemes, and the contributions of transmembrane and amphipathic
helices to the quinone binding sites are much as predicted from model studies.
The present coordinates from the Xia et al. (1) complex from beef heart
mitochondria are from diffraction data to 2.8 Å. A native data set,
has been deposited with the Brookhaven Protein Data Bank as file 1qcr,
due for release July 1998. Data sets with inhibitors (antimycin, myxothiazol,
stigmatellin or UHDBT) bound have been solved, but not yet deposited. The
data from Zhang et al. (2) are from the chicken heart mitochondrial complex
at 3.0 Å resolution, and coordinates for the native structure, and
the complex co-crystallized with stigmatellin and antimycin, have been
deposited with the Brookhaven PDB, as files 2qcr, for release in September
1998, and 1bcc 3bcc, for release in September 1998. Additional structures
for the chicken complex in co-crystals with stigmatellin alone, myxothiazol,
antimycin alone, UHDBT, and several MOA-type inhibitors are also under
refinement. Berry's group have also solved structures for complexes from
the heart mitochonria of rabbit, and two different complexes from beef,
at slightly lower resolution.
Inhibitor binding
The antimycin binding site is close to a b-heme, assigned as cyt bH
on the basis of biophysic evidence. From the Xia et al. work (1), the difference
electron density map ± antimycin showed a strongly defined density
for the inhibitor, and a loss of density which probably corresponds to
a displaced ubiquinone. Zhang et al. (2) modeled density in the native
structure as quinone at the Qi-site, and this density was lost
in antimycin co-crystals.
The Qo-site shows a bifurcated binding pocket (1, 2). The
UHDBT (or stigmatellin) binding domain is close to the 2Fe.2S center in
a lobe distal from cyt bL. The myxothiazol (or MOA-type inhibitors)
binding site is further away from the ISP binding interface, in a lobe
proximal to heme bL. The tails of the inhibitors reach out into
the putative lipid domain, though a relatively narrow orifice which has
a cross-sectional area similar to the tails (3). As a consequence, the
binding domains overlap, and the site would not be expected to accommodate
both types of inhibitor at the same time. This provides a structural basis
for the earlier observation from biophysical studies that occupation by
UHDBT and myxothiazol, or stigmatellin and myxothiazol, is mutually exclusive.
In the Zhang et al. work (2), a weak electron density at the Qo-site
in the native structure may represent weakly bound quinone. The strong
occupancy expected at the site from the double-occupancy hypothesis of
Ding and Dutton (4) is not found in any of the structures from either group.
Crofts et al. (3) have suggested that the two lobes of the Qo-site
are occupied by different intermediates of the quinl oxidation reaction.
The suggestion is based on the bifurcated nature of the Qo-site,
the failure to detect a strongly binding quinone, the different domains
of the two types of inhibitor, and the differential effects of mutation
on inhibitor binding and the interaction of quinone with the ISP detected
through the gx=1.800 EPR band. They suggest that the quinol
bound in the distal lobe forms a reaction intermediate with ISPox,
which leads to electron transfer to form the semiquinone, and that the
semiquinone must move to the proximal site before oxidation by cyt bL.
Arrangement of the prosthetic groups - based on the Zhang et al. structure
of the chicken heart mitochondrial complex, with antimycin and stigmatellin
(PDB file 3bcc).
Colors are:
Monomer 1: cyt bH, green-blue, (top) and cyt bL,
dark green; FeS, orange; cyt c1, yellow
Monomer 2: cyt bH, blue, (top) and cyt bL, dark
blue; FeS, red; cyt c1, pale blue
Both monomers: antimycin, cyan; stigmatellin, magenta.
Note that the head group of ISP from monomer 1 (containing the orange
FeS center) interacts with the Qo-site associated with cyt bL
(blue), and with cyt c1 (paleblue), of monomer 2, and vice versa.
You will need Netscape 3.0 or higher, and Chemscape Chime 1.0 or higher,
to see the model within your viewer
You can download Chime by clicking here.  Chime plug-in
Chime plug-in
Other structural studies
Smith, Cramer and colleagues (5) have previously reported a high resolution
structure for a solubilized cyt f of the b6f-complex from higher
plants, and Berry et al. (6) have reported a similar structure from Chlamydomonas.
A low resolution structure for the complete complex has been reported by
Mosser et al. (7). Iwata, Michel, Link and colleagues have recently reported
on a high resolution structure
for the solubilized Rieske FeS protein from the beef heart complex
(8), and Carrell et al. for the chloroplast Rieske protein (9).
Structure references
-
Di Xia, Chang-An Yu, Hoeon Kim, Jia-Zhi Xia, Anatoly M. Kachurin, Li Zhang,
Linda Yu, Johann Deisenhofer. Crystal Structure of the Cytochrome bc1
Complex from Bovine Heart Mitochondria. Full
text, or Abstract.
-
Z. Zhang, L. Huang, V. M. Shulmeister, Y.-I. Chi, K.-K. Kim, L.-W. Hung,
A. R. Crofts, E. A. Berry & S.-H. Kim. Electron transfer by domain
movement in cytochrome bc1 Nature 392, 677 (1998).
Abstract.
-
Crofts, A.R., Barquera, B., Gennis, R.B., Kuras, R., Guergova-Kuras, M.
and Berry, E.A. Mechanistic aspects of the Qo-site of the
bc1-complex as revealed by mutagenesis studies, and the crystallographic
structure. (Crofts, Berry et al.) Abstract
or Full
text.
-
Ding, H., Daldal, F. and Dutton, P.L. (1995) Biochemistry 34, 15997-16003
-
Martinez, S.E., Huang, D., Szczepaniak, A., Cramer, W.A. and Smith, J.L.(1994)
Crystal structure of the chloroplast cytochrome f refeals a novel cytochrome
fold and unexpected heme ligation. Structure 2, 95-105).
-
Berry, E.A., Huang, L.-S., Chi, Y., Zhang, Z., Malkin, R. and Fernandez-Velasco,
J.G. (1997) Biophysical J. 72, Abstr A125.
-
Mosser, G., Breyton, C., Olofsson, A., Popot, J.-L. and Rigaud, J.-L. (1997)
Projection map of cytochrome b6f complex at 8 Å resolution.
J. Biol. Chem. 272, 20263-20268.
-
Iwata, S., Saynovits, M., Link, T.A. and Michel, H. (1996) Structure of
a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine
heart mitochondrial cytochrome bc1 complex determined by MAD
phasing at 1.5Å resolution. Structure 4, 567-579.
-
Carrell, C.J., Zhang, H.M., Cramer, W.A. and Smith, J,L. (1998) Biological
identity and diversity in photosyntheris and respiration - structure of
the lumen-side of the chloroplast Rieske protein. Structure, 5(12), 1613-1625.
-
So Iwata, Joong W. Lee, Kengo Okada, John Kyongwon Lee, Momi Iwata, Bjarne
Rasmussen, Thomas A. Link, S. Ramaswamy, Bing K. Jap (1998) Complete
Structure of the 11-Subunit Bovine Mitochondrial Cytochrome bc1
Complex. Abstract
or Full
TextScience, 281, 64-71
View of a model structure for cytochrome b from Rb. sphaeroides
The crystallographic structures have provided a nice validation of structural
models derived from sequence analysis, mutational studies, characterization
of the electrogenic reactions of te complex, and biophysical models of
the mechanism. View a model
of cytochrome b from this earlier work in Chime.
The bc1-complex home page
A comprehesive set of information can be found at the
bc-complex site, and the various links available from those pages.
©Copyright
1996, Antony Crofts, University of Illinois
at Urbana-Champaign, a-crofts@uiuc.edu

 Click thumbnail for larger version
Click thumbnail for larger version