NADH:ubiquinone oxidoreductaseComplex I |
The NADH:ubiquinone oxidoreductase (Complex I), provides the input to the respiratory chain from the NAD-linked dehydrogenases of the citric acid cycle. The complex couples the oxidation of NADH and the reduction of ubiquinone, to the generation of a proton gradient which is then used for ATP synthesis. The complex occurs in the mitochondria of eukaryotes and in the plasma membranes of purple photosynthetic bacteria, and the closely related respiratory bacteria. The close homology of sequences, function, and prosthetic groups shows a common ancestry. Mutations in this complex are associated with LEBER HEREDITARY OPTIC NEUROPATHY, MELAS SYNDROME, and possibly with ALTZHEIMERS DISEASE.
A closely related set of sequences is found in chloroplasts; genes for 11 of the 14 minimal subunits are found in the plastid DNA of plants and in the genome of cyanobacteria. However, genes encoding subunits of the NADH dehydrogenase part of complex I are apparently missing in these species, so the complex might lack the NADH processing subunits. It is speculated that the chloroplast enzyme might use the quinone reductase function of the complex with a different reductant,- perhaps ferredoxin or NADPH.


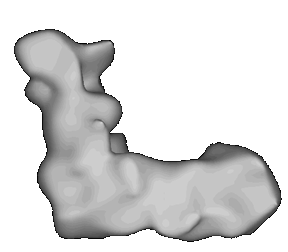
A model of the complex generated by image reconstruction. Click here to see a movie showing the complex rotating.
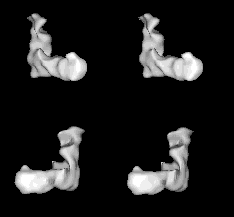
Images from the movie selected to provide stereo pairs for crossed-eye viewing. The two pairs provide approximately orthogonal views.
The images and movie are taken from Dr. Guénebaut's N. crassa complex I page (see ref. below).
Ref.
Guénebaut, V., Vincentelli, R., Mills, D., Weiss, H. & Leonard, K. (1997) Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction. J. Mol. Biol. (1997), 265, 409-418
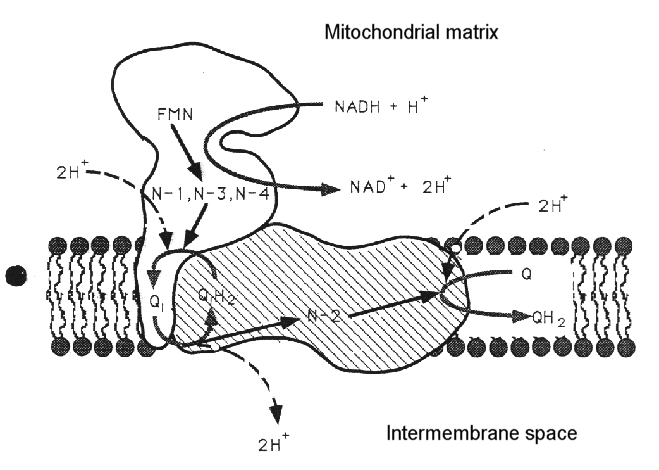
The complex can be dissociated into two main sub-complexes, corresponding to the "ankle" of the boot, and the "foot" of the boot. The ankle is thought to protrude from the membrane so as to be predominantly in the aqueous phase (the matrix side of the mitochondrial membrane, - the N-phase (protochemically negative)), and contains the binding site for NAD(H), and the input electron transfer chain. This consists of a flavine mononucleotide (FMN) prosthetic group as the first acceptor of electrons from NADH, and iron sulfur centers N-1, N-3 and N-4. The sub-complex can be further dissociated into a flavoprotein and an iron protein. The foot (the hydrophobic protein) is membrane bound, and contains a catalytic site at which ubiquinone is reduced, and inhibitors bind, and several iron sulfur centers. There is a second catalytic site for ubiquinone reaction on the ankle, but this is seen as a separate activity only in the dissociated complex.
Subunit Mr Redox centers Function Mitochondria encoded (mainly hydrophobic fragment) ND1 36 Rotenone binding site (possibly UQ reductase site) ND2 39 ND3 13.2 ND4 51.6 ND4L 10.7 ND5 66.9 ND6 18.7 Iron protein(?) Nuclear encoded A subunit 13 Iron protein B subunit 13 23KD subunit 23 N-1b 2Fe.2S(?) Flavoprotein (2 FeS center look like 4Fe.4S from sequences) 42KD subunit 42 49KD subunit 49 N-4 (4Fe.4S) Iron pprotein 51KD subunit 51 FMN, N-3 (4Fe.4S) Flavoprotein, NAD-binding AGGG subunit 12.3
Hofhaus, G., Weiss, H. and Leonard, K. (1991): Electron microscopic analysis of the peripheral and the membrane parts of mitochondrial NADH dehydrogenase (Complex I). J. Mol. Biol. 221, 1027-1043.
Friedrich, T., Steinmüller, K. & Weiss, H. (1995) The proton-pumping respiratory complex I of bacteria and mitochondria and is homologue of chloroplasts. FEBS Lett. (Minireview), 367, 107-111.
Yagi, T. (1993) Biochim. Biophys. Acta 1141, 1-17.
Ohnishi, T., ed. (1993) J. Bioenerg. Biomembr. 25, 325-391.
Dimroth, P. (1997) Biochim. Biophys. Acta 1318, 11-51.
Brandt, U. (1997) Biochim. Biophys. Acta 1318, 79-91.