Lecture 12Proton conduction, stoichiometry |
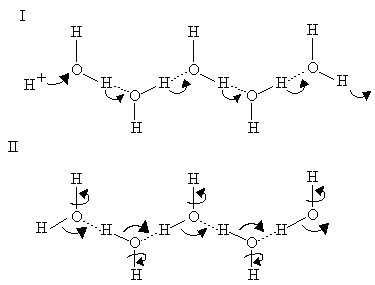
In the "hop" part of the mechanism, a proton first hops from the end of the H-bonded chain to an adjacent group (I, right); transfer of H-bond strength then allows it to be replaced by a H+ binding at the other end, to give the structure in II. In the "turn" phase, rotation of the waters as shown in II then restores the starting structure (I). In this H-bonded chain, the waters can in principle be replaced by suitable protein side chains with H-bonding potential.
Click here for a recent Theoretical Study of H+ Translocation along a Model Proton Wire.
Water is often ordered at surfaces, and it has been suggested that conduction at the surface might be a favored path for the proton current in energy coupling. However, it should be borne in mind that the conductivity of a conducting pathway is the product of the specific conductivity and the cross-sectional area; in most cases, the bulk aqueous phase has a much larger cross-sectional area in any given conduction pathway than the surface, and would therefore be the more probable conductor.
In redox reactions involving oxidation or reduction of quinones, the sites at which protons are released or taken up are often buried in the protein, because the quinone has to reach the site from the lipid phase. In other protolytic reactions involving proton pumping, the sites at which protons are bound or released are often buried in the protein; the center at which vectorial work is done on the proton is relatively immobile, and this requires access pathways through the protein. In both cases, the conduction pathway for protons into the protein is thought to involve channels formed by a chain of bound water molecules and protein side chains. A general picture of such a channel, and how it might operate along the lines of the model above (from Nagle and Tristram-Nagle, 1983), is given below:
The best studied proton channel is that of gramicidin,
shown here in a Chime tutorial. Click here for a free-running
tutorial showing structural features. Eric Jakobsson has simulated the waters in the gramicidin channel using a molecular dynamics in a membrane-gramicidin model.
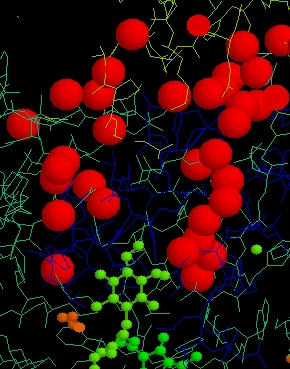
Other nice examples of such a channel have been found in the bacterial
reaction centers from both Rb. sphaeroides and Rps. viridis;
the picture below is of the water channel connecting the quinol at the
QB-site to the aqueous phase from which H+-binding
occurs on reduction of quinone.
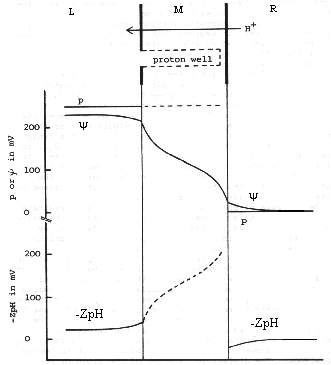
A corollary of the high conductivity of the proton channel is of importance for mechanism. If such a channel is to be part of a proton pumping device, then any work performed on (or by) the proton must be done across some part of the reaction pathway other than the conducting channel(s). To do work on the proton, it must be liganded or sequestered in the "active" part of the pathway.
Proton channels linked to proton "activating" devices are well characterized in two systems in which the prosthetic groups involved have a restricted mobility. One of these systems is cytochrome oxidase, where the work is performed on the proton by coupling redox reactions at the binuclear center to changes in affinity of protein side cahins. Michel and colleagues have suggested a mechanism involving the histidine ligands to CuB. The other is bacteriorhodopsin, where the mechanism of the photocycle has been characterized in some detail, with specific modelling of the reactions by which the Schiff base formed by the retinaldehyde-lysine undergoes deprotonation and protonation during the photocycle.