Succinate:ubiquinone oxidoreductaseComplex II
|
Function
Membrane bound complex which catalyses the following reaction:
succinate + ubiquinone <==> fumarate + ubiquinol
The oxidation of succinate to fumarate is one of the steps in the TCA cycle.
No proton translocation is associated with this reaction.
Subunit composition
Subunit Mr(kD) Prosthetic Groups Function
SDH1 67.1 FAD flavoprotein; succinate binding site
SDH2 28 3 FeS centers iron protein
SDH3 20 cyt b-560 heme membrane anchor for SDH1
SDH4 16.6 none membrane anchor for SDH2
Electron transfer pathway
The catalytic site at which succinate is oxidized is on the flavoprotein subunit (SDH1). The bound FAD acts as electron acceptor. The flavin semiquinone, and the FeS centers in the iron protein (SDH2) act to transfer electrons one at a time to the ubiquinone reductase site. Cytochrome b-560 in SDH3 is not present in all succinate:UQ oxidoreductases, so may not form an essential part of the electron transfer chain.
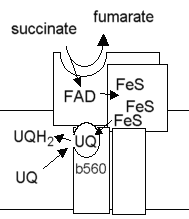
Scheme showing the electron transfer pathway
Fumarate reductase is a similar enzyme which catalyses the reverse reaction, but using menaquinol as electron donor in some bacteria. Two bacterial structures have been solved, for enzymes from E. coli and Wolinella succinogenes. E. coli is from the g-proteobacteria, while Wolinella succinogenes is from the e-proteobacter, with the latter as more deeply branched in phyllogenetic trees. Although the two proteins show the same basic structure, and have obviously evolved from a common ancestor, the structures show several quite different features. The enzyme from E. coli contains two menaquinones bound in a heme-free membrane spanning part containing two subunits, while the enzyme from Wolinella succinogenes shows two hemes, and no bound quinone, in a single subunit membrane spanning part (see ref. 1 for a discussion of the evolution of these spans).
Click here for a Chime tutorial on E. coli fumarate reductase (ref. 2)
Click here for a Chime tutorial on Wolinella succinogenes fumarate reductase (ref. 3)
Yeast mitochondrial enzyme
Summary of prosthetic groups, function and sequence information for the yeast mitochondrial enzyme.
- SDH1. Succinate dehydrogenase (ubiquinone) flavoprotein (Fp) subunit, with FAD binding domain.
- SDH2. Succinate dehydrogenase (ubiquinone) iron-sulfur protein (Ip) subunit, contains 3 FeS centers.
- SDH3. Succinate dehydrogenase membrane anchor subunit for Sdh1p. Cytochrome b-560 binding subunit.
- SDH4. Succinate dehydrogenase membrane anchor subunit for Sdh2p.
References
- Hägerhäll, C. and Hederstedt, L. (1996) A structural model for the membrane-integral domain 0d succinate:quinone oxidoreductases. FEBS Lett. 389, 25-31
- Iverson, T. M., Luna-Chavez, C., Cecchini, G., Rees, D. C. (1999) Structure of the E. coli Fumarate Reductase Respiratory Complex. Science 284, 1961
- Lancaster, C.R.D., Kroger, A., Aver, M. and Michel, H. (1999) Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature 402, 377 - 385
©Copyright 1996,
Antony Crofts, University of Illinois at Urbana-Champaign,
a-crofts@uiuc.edu
