








|
Mammalian Reovirus Core
as solved by Cryoelectron Microscopy and Image Reconstuction
Rotating around y axis
 MPEG version (989K)
MPEG version (989K)
 Quicktime version (487K)
Quicktime version (487K)
Cropping frontal sections
 MPEG version (120K)
MPEG version (120K)
 Quicktime version (164K)
Quicktime version (164K)
Cropping radial sections
 MPEG version (164K)
MPEG version (164K)
 Quicktime version (236K)
Quicktime version (236K)
Summary
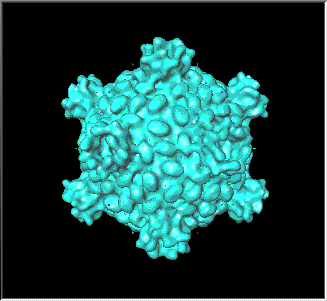
Animations of the core particle of mammalian reovirus, as solved by cryo-electron
microscopy and image reconstruction. Rendered as an isosurface
using Iris Explorer
on a Silicon Graphics workstation.
Isosurfaces are used to portray the three-dimensional structure of the
virus. An isosurface is obtained by converting the subset of electron
density values in the three-dimensional array that contours a chosen
threshold value (isovalue) into a surface representation approximated
by polygons.
Structural elements of interest include the "turrets" at the five-fold
vertices containing lambda-2 protein and the nodules on the remaining
surface. Note that the mu-1 trimers and the sigma-1 spike protein are
lost in the transition from ISVP to core.
Created October 1994.
Acknowledgements
We gratefully acknowledge Timothy S. Baker and colleagues at the
Structural Biology Group at Purdue University for supplying
us with the three-dimensional cryo-electron microscopy datasets
which were used to generate these images and animations.
Reference
Dryden, K.A., G. Wang, M. Yeager, M.L. Nibert, K.M. Coombs,
D.B. Furlong, B.N. Fields, and T.S. Baker. 1993. Early steps in reovirus
infection are associated with dramatic changes in supramolecular
structure and protein conformation: analysis of virions and
subviral particles by cryoelectron microscopy and image reconstruction.
J. Cell Biol. 122:1023-1041.
 Go to the
top page Go to the
top page
© 1994 Stephan Spencer. Institute for Molecular Virology/
sspencer@netconcepts.com
 |

