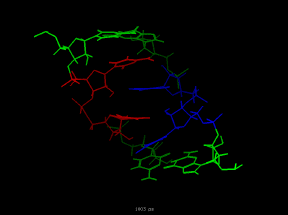
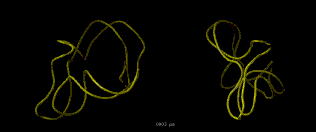
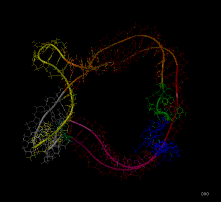Poliovirus 3' UTR RNA dynamics were calculated according to Hilbert's description in
calculations.html.
The original data FLI movie files which were converted to
standard QuickTime movies, as well as 30% and 50%
reduction QuickTime and animated GIF movie files (by Jean-Yves Sgro).
The table below allows you to see the 30% animated GIF version.
QuickTime versions are available here.
- RNA Dynamics -
--- click on images to see animations ---
bulge-solid
| bulge-stick
|

| 
| renderall
| rotatex
|
| 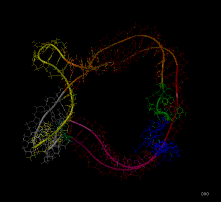
| |
- Wang J, Bakkers JM, Galama JM, Bruins Slot HJ, Pilipenko EV, Agol VI, Melchers WJ
Structural requirements of the higher order RNA kissing element in the enteroviral 3'UTR.
Nucleic Acids Res 1999 Jan 15;27(2):485-90
- Melchers WJ, Hoenderop JG, Bruins Slot HJ, Pleij CW, Pilipenko EV, Agol VI, Galama JM
Kissing of the two predominant hairpin loops in the coxsackie B virus
3' untranslated region is the essential structural feature of the
origin of replication required for negative-strand RNA synthesis.
J Virol 1997 Jan;71(1):686-96