Chromosomes and genes can alter as a result of structural changes. These changes are called mutations. The original state can usually not be re-established. The loss of hereditary information is irreversible. A change caused by a mutation is kept throughout all following generations, if it does not cause lethality. A number of different mutations were identified with the help of polytene chromosomes. They occur also in normal chromosomes.
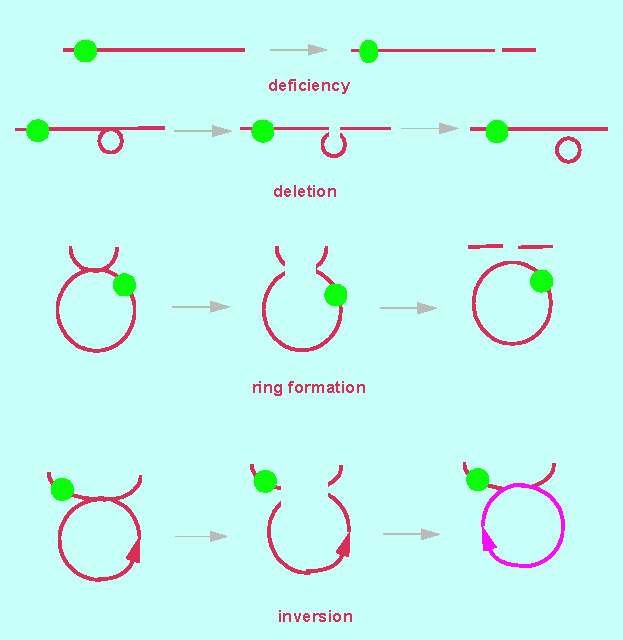
A deficiency is the loss of a chromosomal end fragment. Since it occurs normally only in one of two homologous partners, the result is a pairing of a defect and an intact chromosome. Deficiencies can be recognized by this. The intact partner chromosome shows how large the missing fragment of the defect chromosome is.
A deletion is the loss of an inner chromosomal fragment. It can be recognized by a loop of the intact partner chromosome at the site where the missing fragment would have been in the defect chromosome. The loop allows conclusions about the size and the site of the deletion.
A duplication is the doubling of one or several chromosomal fragments. It shows also as a loop but this time, the loop is in the defect partner chromosome. To find out, whether the loop has been caused by a deletion or a duplication, the band pattern of the chromosome has to be analyzed. The mutation is caused by a duplication, if the band pattern of the loop occurs twice. Usually, the pattern can be found again in a domain neighbouring the loop. The loop is caused by a deletion, if a fragment is missing.
An inversion is the change of direction of a chromosomal segment. It results from a segment that has broken out of the chromosome and fused again at the same site but with inverted direction. A pairing of homologous chromosomes is possible, though rather complex loops may be formed.
How do chromosomal mutations take place ? The deficiencies are easiest to explain. They are the result of a simple chromosomal fraction. Numerous mutagens, like chemicals or ionizing radiations lead to an increase in the probability of such fractions. Chromosomal fragments without a centromere are lost during the following mitosis. They are not integrated into one of the two daughter nuclei. Usually, they remain in the equatorial plane, where they are degraded.
All the other chromosomal mutations can be explained by two subsequent events: fraction and fusion. Crossing-overs are caused by the same mechanism. In a strictly formal sense, deletions and duplications can thus be described as 'illegitimate' crossing-overs. A 'normal' crossing-over results in two chromatids that are homologous to the original ones (i.e. of the same length and with the same number and order of gene loci). 'Illegitimate' crossing-overs do in contrast result in two products of unequal length. As a consequence, one chromatid loses a fragment (a deletion) to its partner chromatid. This does now harbour the fragment twice (a duplication). Usually, duplicated fragments are arranged in a tandem.
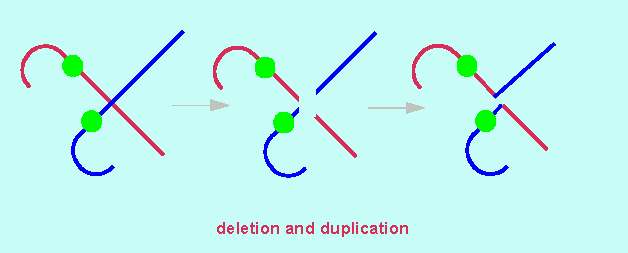
The classic example for the way in which a deletion/duplication
event takes place is the Bar-gene of Drosophila (the
Bar-locus; Bar is written with a capital B due to its
dominance over the wild type). The eyes of Drosophila are
normally roughly oval. But those of the Bar-mutant are shaped
like a bar. This phenotype is caused by a duplication. The
Bar-locus can be traced back to a certain chromosomal band.
'Illegitimate' crossing-overs that occur in this domain can on one
hand restore the wild-type state, on the other hand they do create a
domain in the homologous chromatid, where the Bar-locus occurs
three times, one behind the other (double Bar).
The expression of the characteristic Bar shows that it is not only the lack or the defect of a gene that can cause a change of the phenotype, but also the surplus of an additional locus. In a normal, diploid genome, two alleles exist for every gene. They are on separated, but homologous chromosomes and they are therefore said to be in trans-conformation. After a duplication, the respective gene occurs in a diploid genome for four times. The intact chromosome harbours the gene two times, once on each of its two chromatids, a trans-conformation. The homologous mutated chromosome has also two copies of the gene, but one chromatid has no copy, while the other one has two (a cis-conformation):
C and C are also called
pseudoalleles. They are not
separated during meiosis, unless a crossing-over takes place. An
allele and its respective pseudoallele may accumulate mutations
independently and thus go through autonomous evolutionary
processes. The effects of an allele and its pseudoallele are only
rarely additive. A pseudoallele does thus not replace an allele. This
and other observations lead to the discovery that the genes of a
genome are no independent units. Their activities are actually
controlled by neighbouring DNA domains. This phenomenon was
originally called the position
effect. Molecular analyses begin to elucidate how such
control mechanisms of single genes work.
The formation of an inversion was first analyzed
by B. McCLINTOCK (1938) in maize chromosomes . After the fraction,
two homologous fragments join so
that one chromosome without a centromere forms, while the other
chromosome has two. The chromosome without a centromere is lost.
During anaphase, the two centromeres of the other chromosome
segregate to opposite poles, thus causing a break of the in-between
domain. Since the exact place of the breaking is subjected to chance,
one of the daughter cells will get an elongated, the other one a
shortened fragment. In the elongated one, the additional domain is
inverted.
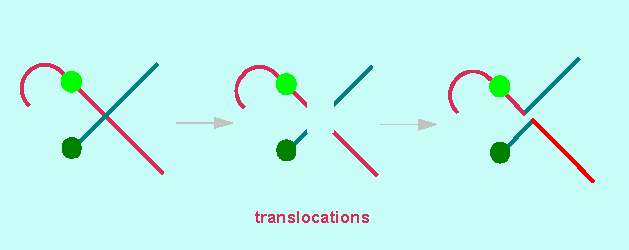
Translocations occur as a result of the pairing of non-homologous chromosomes during meiosis. The classic example is Oenothera. It has already been said that Oenothera lamarckiana (Evening Primrose) can only survive in its hybrid form. In contrast to many other animal and plant species, it is impossible to correlate the linkage groups and the chromosomes of Oenothera. There exist n = 7 chromosomes but just one coupling group (= complex, either gaudens or velans).
The cause is a series of subsequent translocations that were analyzed by R. E. CLELAND (University of Indiana, Bloomington, 1949). CLELAND symbolized the information of each chromosome by two numbers, separated by a point:
This is contrasted by the paternal set of chromosomes:
During meiosis, the homologous maternal and paternal domains pair. Normally, these domains and homologous chromosomes are identical. This is different here. Due to the regular, balanced translocations, the chromosomes of Oenothera and a few other species form a ring when pairing: domain 2 of the maternal chromosome 1.2 pairs with domain 2 of the paternal chromosome 2.3. This again pairs with chromosome 3.4 and so on. The ring is closed by the pairing of chromosome 14.1 with chromosome 1.2. During anaphase I, the centromeres are distributed onto the daughter cells in a strictly alternating way. The result is that the chromosomes of the original maternal set stay together in one daughter cell and those of the paternal in the other. This explains the occurrence of just one coupling group.
The system works usually very well, though it is more prone to disruptions than normal meiosis is, leading to an increased mutation rate in Oenothera. This may be the reason, why H. de VRIES detected the phenomenon mutation in just this genus. Similar conditions were found in Rhoeo discolor, Paeonia californica and in species of the genus Datura.
|
|