|
|
|
Friedrich Miescher was born in Basel on August 13th, 1844. In 1869, he discovered the nucleic acids, the molecular substrate of the genetic code. The work was done in the laboratory of Felix HOPPE-SEYLER, which was located in the castle of Tübingen. He also demonstrated that the regulation of breathing depends on the CO2 -concentration in the blood. In 1872, he became a Professor at the University of Basel. He died in Davos on the August 26th, 1895.
Nucleic acids are polymers consisting of nucleotides that are linked by phosphodiester bonds (polynucleotides). Depending on the type of sugar present in the nuclotide (ribose or desoxyribose), it is distinguished between two classes of nucleic acids:
![]() ribonucleic
acids (RNA) and
ribonucleic
acids (RNA) and
![]() desoxyribonucleic
acids (DNA)
desoxyribonucleic
acids (DNA)
DNA is known to be the carrier of genetic information, RNA functions as a messenger (messenger RNA) and takes part in protein synthesis (transferRNA, ribosomalRNA). As in proteins, the linkage of the monomers to polynucleotides is directed, though the process is no condensation but a polymerization where a pyrophosphate residue is split off.
To distinguish the atoms of the bases and those of the sugars from each other, the latter are marked by a '. It is thus spoken of 1', 2'.... The polynucleotide chain grows (during synthesis) from the 5' to the 3' terminus. Written this way, the sequence of nucleotides is colinear to the sequence of amino acids in a protein (from N- to C-terminus; see also genetic code).
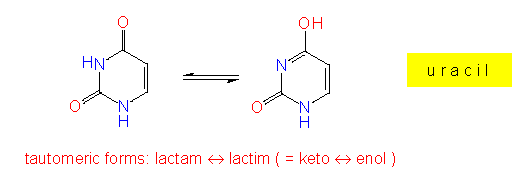
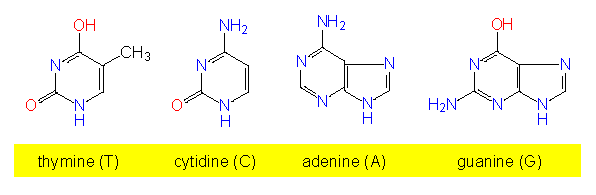
Besides their sugars, RNA and DNA do also differ partially in the bases. RNA contains usually purine and pyrimidine bases, while DNA contains thymine instead of uracil. Especially in RNA but also in DNA, so-called rare, modified bases occur. Some of them are intermediate products of the normal pyrimidine or purine synthesis.
|
|
The sequence of nucleotides within a nucleic acid may seem arbitrary at first, but we know that it is this sequence that encodes the genetic information. Though not every nucleotide sequence contains information. Long, repetitive DNA sequences exist and their function is not known. Since around 1975, methods for the sequencing of nucleotides have been developed.
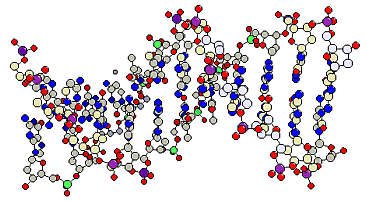
DNA contains A and T or C and G respectively in equal amounts. The ratio A+T/C+G is specific for every species. Based on these data and Röntgen structure analysis, J.D.WATSON and F.H.CRICK did develop the WATSON-CRICK-model in 1953. Its chief property: it explains the mechanism of the genetic material's replication by using both of the DNA strands as matrices. The further properties of the model are:
|
|
Both polynucleotide chains are wound around each other in an antiparallel way. |
|
|
The purine and pyrimidine bases are directed towards the inner of the helix, the phosphates and the sugar residues are at the outside. The planes of the bases are at a right angle to the axis of the helix. |
|
|
The radius of the helix is 10Å, the distance between neighbouring bases is 3.3Å. |
|
|
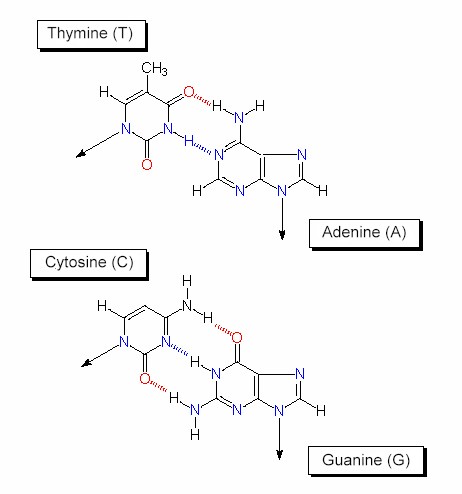
Both chains are linked by hydrogen bonds that develop between a pyrimidine and a purine base. An adenine is thus always linked with a thymine, a guanine with a cytosine. Between adenine and thymine exist two hydrogen bonds, between cytosine and guanine three. |
|
|
The specific base pairing causes complementarity. If the sequence of one strand is known, then that of the other can be predicted. |
The mechanism of replication was elucidated in 1958 by M.MESELSON and F.W.STAHL, who could experimentally prove a prediction made by WATSON and CRICK and thus showed that DNA duplicated in a semiconservative way.
In contrast to DNA, RNA is normally single-stranded. But it displays also some double-stranded, helical sections that are caused by folding of the single strand. These sequences are mirror images of each other and develop palindromes.
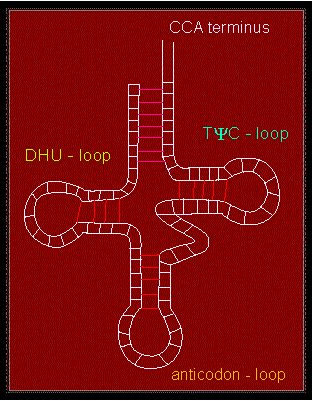
The clover-leaf structure of tRNA, a group of roughly 80 nucleotide long molecules that is needed for protein synthesis is known. They function as adapters. RNA contains a number of rare and modified bases that develop additional, exceptional hydrogen bonds and do thus stabilize the specific tertiary structure.
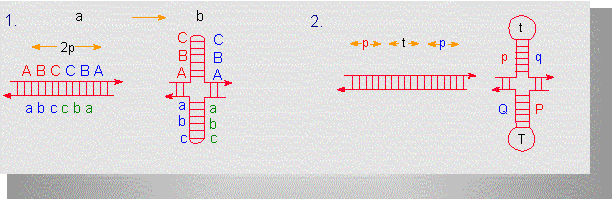
Two types of inverted (mirror images) repetition units. Each unit consists of 2p nucleotides that have to be connected via hydrogen bonds to be able to form a palindrome (b). In (1), the units are separated by non-repetitive sequences. In (2), this is not the case. The sequence is t nucleotides long. Please notice that a few bases at the vertex of the palindrome stay unpaired (according to D. A. HAMER, C. A. THOMAS, 1974).
Complex forms of secondary structures can be
found in ribosomal RNA
(rRNA). Its elucidation during the
last years gave valuable proof for the
relationship between the rRNA of higher plants
and that of cyanophyta and was thus an important
pillar for the endosymbiont theory. On the other
hand, such analyses do also proof the existence
of three lines of
development of cellular organisms:
archaebacteria, eubacteria and the line that
lead to the development of eukaryotes.
|
|