Die hier wiedergegebene Abbildung ist ein Screenshot der interaktiven Karte der KEGG: Kyoto Encyclopedia of Genes and Genomes. An diesem Beispiel wird demonstriert, wie die KEGG-Datenbank aufgebaut ist. In Form eines INTERFACE zu ihr wird gezeigt, wie man einen vollständigen Zugang zu den Daten dieser Datenbank - d.h. zu sämtlichen Angaben über die am Stoffwechsel beteiligten Enzyme sowie den Intermediär- und Endprodukten erhalten kann.
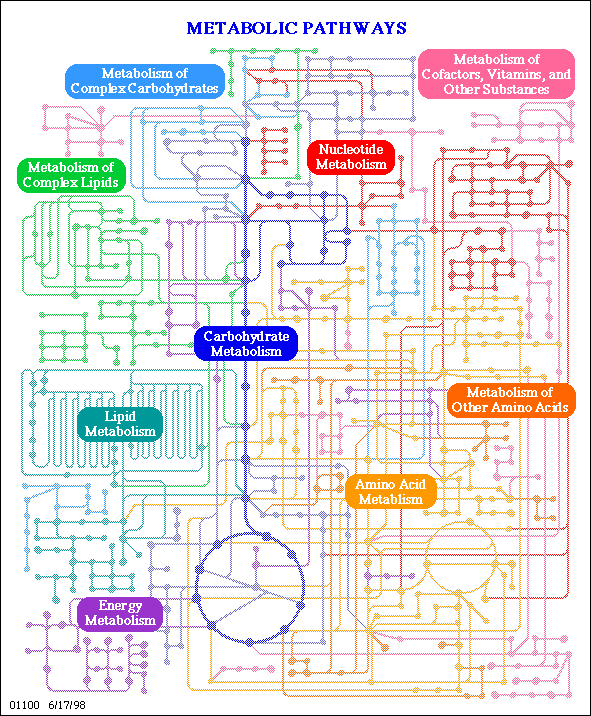
Wie arbeitet man mit der Karte? - ein Beispiel: Durch Anklicken von Aminoacid Metabolism erhält man einen Kartenausschnitt.
Die nachfolgenden Listen dienen dazu, einen systematischen Zugang zu allen Datenbankeintragungen von KEGG - Metabolic Pathways - zu erhalten (alle Links führen direkt zur Datenbank in Kyoto/Japan!).
| KEGG Metabolic PathwaysGraphical pathway maps and ortholog group tables |
|---|
Note - The ortholog group table contains only a subset of enzymes in the pathway map. The subset represents a correlated cluster of enzyme genes that appear in a localized area in the genome and that function at closely related positions in the reaction pathway.
[ KEGG Regulatory Pathways | KEGG Table of Contents ]
Last updated: August 11, 1998
Peter v. Sengbusch - b-online@botanik.uni-hamburg.de