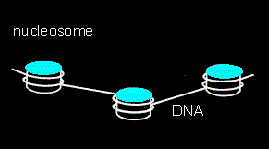
DNA occurs in the nucleus normally complexed with histones. These two structures together form the chromatin. Most eucaryotic cells have five types of histones: H1, H2a, H2b, H3 and H4. A complex of two molecules H2a, H2b, H3 and H4 each forms a nucleosome, a disc-shaped structure. The DNA is wound around it. On average one nucleosome occurs per 200 base pairs. 140 of them are wound around the nucleosome in 1 ¾ coils, the other 60 are free and connect the single nucleosomes. H1 does not belong to the nucleosome complex but serves to enhance the condensation of the chromatin even further.
 |
Until a few years ago it was generally assumed that the histones take part in the selective transcription of single DNA segments (genes or groups of genes). This proved to be wrong: a group of acidic nuclear proteins (non-histones) fulfils this function. Non-histones is a collective term for a number of regulators that are acidic when compared to the strongly alkaline histones. Otherwise their amino acid composition is within the usual framework. |
Clue exists that certain protein fractions bind to certain DNA segments. Further experiments hint at their possible activation by hormones, metabolites, etc. Additionally it has shown that the phytochrome system transfers signals to the DNA that enhance the transcription of certain segments selectively.
Among the acidic nuclear proteins are also the polymerases that catalyze replication and transcription, as well as proteins that form RNP-complexes (ribonucleoprotein complexes) with the RNA.
The degree of condensation of the nuclear DNA is variable. Strongly condensed DNA is called heterochromatin, less condensed one euchromatin. Heterochromatin is stained more intense than euchromatin by certain nuclear dyes.
Condensation and decondensation of the DNA are at least partially reversible processes. Each DNA segment that has to be replicated or transcribed has to be decondensed during this time. The highest degree of condensation can be found in the chromosomes (mitosis, meiosis) during cell division.
Eucaryotic DNA consists of repetitive ( r ) and non-repetitive ( s ) segments, as has been mentioned before. These segments occur in three different patterns of organization:
r1r1r1......r2r2r2r2.......r3r3r3........rnrnrn
The number of the different repetition units (rn) and the number of repetitions is far larger than 1000. It is also spoken of clusters.
r1r1r1 . . .r2r2. . .s1 . . .r3r3r3. . .s2s3s4r4r4r4. . .rn
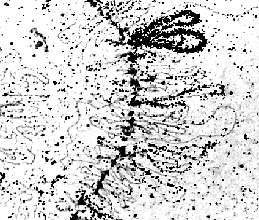
Part of a lamp brush chromosome of Triturus cristatus (an American newt-species): 'intermediate' repetitive DNA is hybridized in situ with [3H]-labelled RNA transcripts at the loops of the lamp brush chromosome. Only few loops are labelled in this way (H. C. MacGREGOR, Leicester, 1977).