

In his final year at school, Mick studied Biology, Anatomy, Physiology and Hygeine and Chemistry.
After finishing school, Mick obtained the degrees BSc, from Glasgow College of Technology, and PhD in Biochemistry from the University of Dundee in Scotland.
Since finishing his PhD, Mick has worked at:
Michael's wife, Anabel, is originally from Uraguay. They have a daughter, Emily who will be three in October. Mick's three sisters and one brother all live in Scotland.
Like most Scots, Mick is a soccer fan. He plays in an indoor soccer competition in a team called the 'Biohazards'! In his spare time, Mick writes short stories. He also loves skiing and watching Clint Eastwood movies.
Interviewer: Can you describe briefly how you are doing whatever you do?
Dr Foley: We use a mouse which has been immunised against malaria as the source of our DNA. We are looking for the genes which code for the antibodies which protect the mouse. When we have a little DNA, we use the technique of PCR to get lots of the same DNA to work with.
We use the DNA to create a 'library' of antibodies attached to the surface of a particular virus. Not all of the antibodies will be useful to us, but we can then select for the antibody we want in the virus pool by binding to the corresponding antigen. If we repeat the process of incubating the viruses with antibody and finding those that bind best, we can get a high quality antibody and the gene for this antibody. This can then be tested for its ability to protect non-immune mice form the malaria parasite.
Interviewer: Who or what (animal or plant) will benefit from your research/techniques?
Dr Foley: People who live in, or visit, the areas of the world where malaria is endemic.
Interviewer: What are the economic implications of your applied genetics? (How expensive is it to do? What will be/are the cost benefits of the outcome?)
Dr Foley: Malaria is prevalent in more than 90 countries in the world, threatening 40 per cent of the world's popluation -- 2400 million people. It is estimated that there are 300-500 million cases of malaria a year and 1.5 - 2.7 million deaths. Most of these deaths are children under 5 years of age.(World Health Organisation, 1996) International expenditure on malaria research is about $84 million, or $42 per death.
By comparison, the international research money spent on HIV/AIDS is about $3274 per death, and for asthma, about $789 per death.
It's sad that a disease which causes such hardship in the world's less wealthy nations is so poorly funded. But if the money spent on malaria research produces a reliable treatment, it will have been money very well spent.
Interviewer: Historically, what is the scientific background to the research you are now doing?
Dr Foley: This project combines three areas of biology which interest me. The areas of virology and parasitology are fascinating in themselves. And it is exciting to be able to use these, together with the techniques of biotechnology to help solve an important question such as this.
Interviewer: For a Year 12 Biology student to understand what you are doing, some background knowledge of biological concepts will be needed. Can you tell me what biology is relevant to what you are doing, and why?
Dr Foley: Many biological concepts are relevant to this research.
Some understanding of the life cycle of the malaria parasite, Plasmodium species, is necessary to understand why this is such a problem in certain countries of the world. (Interviewer: The students would have very likely studied this earlier in the year and could find information about it in their VCE Biology text.) Students also need to know that there is a conserved protein (a protein with the same structure) amongst the different species of Plasmodium. Evolution has presumably retained this protein for some reason.
Students also need to understand the life cycle of the virus we use. It is a bacteriophage, and once we have put the DNA of interest into the virus, we can use it to infect bacteria which will rapidly make millions of new virus particles containing the DNA of interest for us.
To understand how we get the genes for the antibodies we want, students need to uderstand a little immunology. We raise antibodies in mice by injecting them with antigen from the malaria parasite. This leads to antibody formation by plasma B cells. Much of the antibody production goes on in the spleen, so we use the mouse spleen to extract mRNA. The reasoning here is that if the gene for the antibody is being expressed here, there must be plenty of specific mRNA formed in the spleen cells' nuclei by transcription, and taken to the ribosomes for translation into the antibody protein.
Once we have the mRNA we use reverse transcriptase to get cDNA. Students need to understand the relationship between mRNA and DNA to follow this.
At this point we have the cDNA for all of the mRNA molecules produced by the mouse. This means that we have cDNA for antibodies mixed with cDNA for all the structural and functional (eg enzymes) proteins made in the mouse spleen. Separating out the cDNA for antibodies is the first task.
We amplify only those DNA molecules coding for antibodies. We do this using PCR and antibody specific primers. So now we have a cDNA pool for all the antibodies made in the mouse spleen, but we still need to locate the genes for the anti-malaria antibodies from this pool.
We put each of the genes into a virus and let the virus make the antibody protein on its surface. The virus is shown in the following diagram. You can see on the diagram how the virus has 'arms' and the antibody protein is displayed on one of those arms.
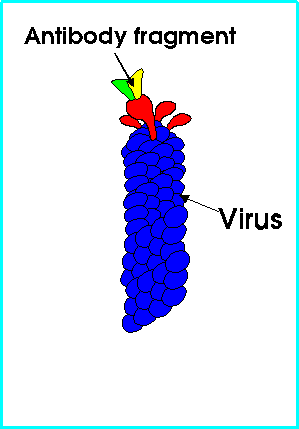
Next we use antigen-antibody binding to select for the desired antibodies (those against our malaria protein). This process gives us viruses containing the genes we want, as demonstrated by their ability to make the antibodies which bind to the antigens.
Once we have the DNA which codes for the antibodies we are seeking, we try to cause mutations in it. We do this because in producing a variety of antibodies we are hoping to improve on nature's antibodies originally made in the mouse.
Interviewer: I now need to know something of the techniques used in your research. Can you tell me what techniques are important, and why they are important?
Dr Foley: A flow chart shows what we do in a simplified form:
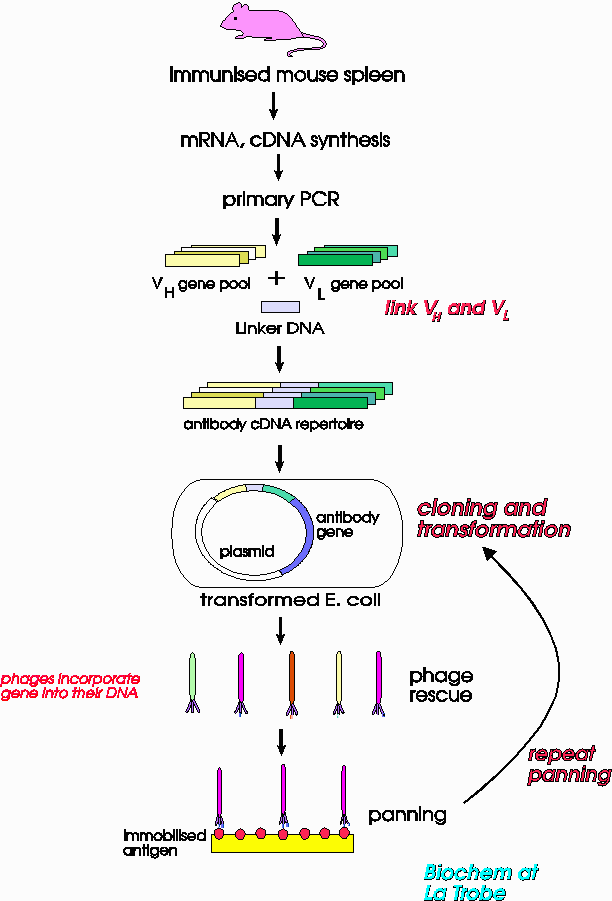
Then we use chemical methods to isolate the mRNA from the mouse's spleen.
We use the enzyme reverse transcriptase to produce cDNA from the mRNA. Then we use PCR to amplify this cDNA - basically, we need to get enough cDNA to work with from the small amount we obtain initially.
Because antibodies are proteins which contain two chains linked together we get the cDNAs for all possible versions of these chains. We call the chains the light, VL (because they are shorter)and the heavy chains The cDNA which codes for antibodies is isolated from the mixture of DNAs and these genes are inserted into viruses. To do this, we first put the genes into plasmids, using restriction enzymes to cut the DNA and create 'stick ends', and DNA ligase to join the pieces of DNA up again into plasmids. The plasmids are inserted into E.coli, which are then incubated with the virus. The virus is actually a filamentous phage named M13. As is normal, the viruses replicate in the E.coli, but some of the transgenic plasmid DNA becomes incorporated in the new phages. We call this 'phage rescue' since the phages 'rescue' our DNA from the E.coli. The viruses now contain all of the many different cDNAs which cause them to express the antibody proteins on their surfaces as shown in the earlier diagram.
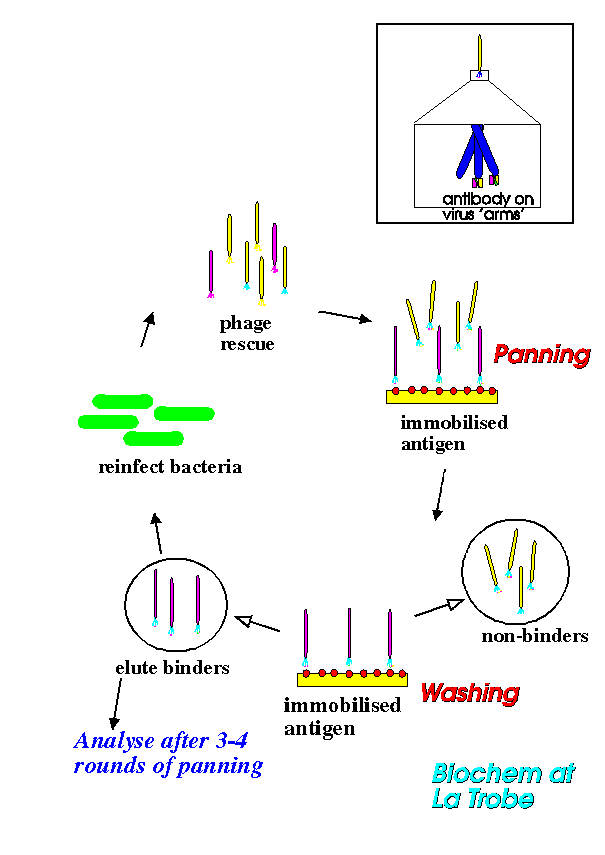
To find the viruses with the antibody we are seeking, we need to put the virus mixture through a selection process called panning. As the name suggests, this is a bit like 'panning' for gold; we are looking for the desired phage amongst many thousands of phages.First we coat a suitable container with the antigen of interest. Then we add the virus mixture to the container, in a suitable solution of course. The viruses with the antibody we want bind to the antigen, the others do not bind and remain in solution. The unwanted viruses can be washed out of the container, the wanted viruses remain bound on the sides until we alter the conditions and free them. We can then grow up new batches of these viruses in E.coli and repeat the selection process. After three or four rounds of this we obtain a highly enriched virus preparation ready for further experiments. Essentially, in this process of panning we have purified the gene we are seeking - it just happens that the gene is conveniently packaged in a virus. The diagram below shows this panning technique.
Interviewer: There is often discussion or debate about issues associated with biotechnology. What is one such issue relating to your work? Can you outline the arguments of the opposing sides of the debate please?
Dr Foley: A major issue in malaria research is 'who is going to pay for it?' The large drug companies who are working on other biotechnology research are not interested in this. They are very aware that the countries where a new antimalarial treatment would be most useful are the countries which are least able to pay for it.
On the Internet there is information about research into malaria and other tropical diseases at "Malaria and Tropical Disease Weekly", and students could try searching for 'malaria' within the "multimedia reference library". Also, if you go to the 'search' page at the WHO www site and type 'malaria' you'll get lots of information about the disease and about WHO programmes that deal with it.
Other Internet sources of information are at the Malaria Foundation, and at the Australian Academy of Science NOVA Web site.
Interviewer: Thank you for your time.
Reference: World Health Organisation. WHO facts. (WHO, www site), visited 13 May, 1997
For ideas on where you might find additional resource material, look again at the section Resources on another page at this site. And don't forget the power of the Internet to provide you with information. A well thought out search, using a search engine such as Alta Vista, should provide more information than you can possibly use!
Please do not phone Dr Foley for further information for your CAT. He, like the rest of the staff in Biochemistry, is a busy person. However, if you think we can help you some more, use either the comment form or the email address for this site and we will do what we can.
If you would like to read another interview with a La Trobe Biochemistry scientist, you can click here to go back to the Interview Index.
If you would like to 'netsurf' for some more resources, you might find the author's favourite Biology Bookmarks helpful.