Transcription is the process during which the genetic information is transcribed from DNA to RNA. One of the two DNA strands is used a template for the fabrication of a complementary RNA. The RNA retains all the information of the DNA from which it was copied, as well as the base-pairing properties of the DNA. Transcription is similar to DNA replication in that one of the two DNA strands acts as a template on which the base-pairing abilities of the incoming RNA nucleotide are tested. The RNA nucleotide is only incorporated as a covalently linked unit if a good match with the DNA template is achieved.
But DNA transcription differs also from DNA replication. The transcription product, the RNA, for example, does not remain annealed to the
DNA template. Instead the two strands of the original DNA
helix anneal again directly behind the region where the ribonucleotides
are being added thus replacing the the RNA chain. The newly synthesized
RNA molecules are also very short compared to DNA molecules since
only limited regions of DNA are copied. These regions correspond
to whole proteins or part of a protein. Only in some small viruses
with genomes ranging around three genes the transcription can
occur en bloc.
Transcription is catalyzed by RNA polymerases. The procaryotic enzyme differs from the eucaryotic one in several decisive properties. The transcription products can be grouped into three different classes according to their function:
![]() messenger RNA (mRNA)
messenger RNA (mRNA)
![]() transfer RNA (tRNA) and
transfer RNA (tRNA) and
![]() ribosomal RNA (rRNA).
ribosomal RNA (rRNA).
In eucaryotes not mRNA but a less clearly defined class called hnRNA (heterogeneous nuclear RNA) is synthesized. Its main portion is degraded immediately after synthesis. Only a smaller portion, the product of partial degradation and reorganization is processed to mRNA in a complex and highly specific process called splicing. During splicing the transcript is capped at the 5' end, its introns (non-coding regions) are removed and it is polyadenylated at the 3' end. The whole process takes place within the nucleus and the ready mRNA is finally transported through the nuclear pores into the cytosol where it is available to be translated.

The excision of sections out of an RNA molecule is called gene splicing. To prove whether it has taken place RNA strands are fused with their respective DNA counterparts (which has been molten and thus rendered single-stranded). The mRNA-DNA hybrid shows DNA loops wherever a part of the mRNA has been removed (H. WESTPHAL and S. P. LAI, Bethesda, 1977)
RNA has many functions. Since some years nucleotide sequencing methods are common and many RNA species have been analyzed. The sequences contain information about:
| the folding, i.e. the secondary and tertiary structure of the RNA-molecule, | |
| regions interacting with proteins or other nucleic acids, | |
| the relations between a gene and its transcription product. The comparison shows how the RNA has been processed in order to achieve a final (mature and fully functioning) product from a precursor. | |
| It shows, too, the degree of RNA homology between different species. This information can be quite useful to elucidate the relationship between the respective organisms. |
Extensive picture material of molecular models and atom co-ordinates (macromolecules, small molecules and molecular complexes) can be found at:
(The Image Library of Biological Macromolecules, The RNA World, etc.)
mRNA is the carrier of genetic information, i.e. it contains the instruction for the synthesis of one (in procaryotes often also several) polypeptide chains. The mature RNA of eucaryotes contains between 400 and 4000 nucleotide bases. Beginning and end contain additional sequences added after transcription. Many though not all mRNA molecules have a so-called cap (= capping), a specific oligonucleotide in which the bases are linked in a way otherwise very uncommon in nucleic acids: at the 5'-terminus. And 30 - 40 percent of all mRNA isolated from the cytosol carries a poly-A sequence (= polyadenylation) (up to 200 nucleotides) at the 3'-end. Only part of the mature RNA is translated into a protein. At the beginning of the mRNA, just behind the cap, is a non-coding sequence, the so-called leader sequence (in the range of 10 - 200 nucleotides). The coding sequence starts with the initiator codon AUG and ends with one of the three stop codons (UAG, UAA or UGA).
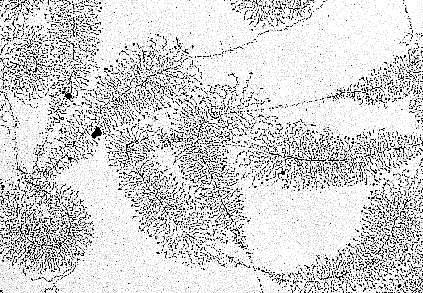
In the nucleoli of the oocyte nucleus of Triturus virescens, an American newt species (as well as with other amphibians) free DNA occurs. The picture shows the transcription of genes that are carriers of the information about ribosomal RNA formation ("MILLER-trees"). More about the interpretation of the electron microscopic picture (O. L. MILLER, B. R. BEATTY, Biology Division, Oak Ridge National Laboratory, 1969).
The leader-sequence is followed by another non-coding sequence of up to 600 nucleotides length. Only rather few plant nuclear acid sequences (genes, non-coding sequences) have been sequenced until now. Known is, for example, the nucleotide sequence of the gene for the small subunit of ribulose-1,5-bisphosphate carboxylase. It is 668 nucleotides long, 440 of which are coding.
tRNAs are a group of rather small molecules. They have a length of 70 - 90 nucleotides many of which are modified and belong to the rare bases. The modifications take place after transcription with the help of specific enzymes. tRNAs fold themselves and form a characteristic secondary structure called cloverleaf structure. A further folding leads to a defined tertiary structure that is stabilized with the help of the rare bases. tRNA has two main functions:
| The recognition of a mRNA codon by base-pairing (codon-anticodon recognition) and | |
| the recognition of the respective amino acid. This second
recognition is helped by a group of enzymes, the aminoacyl-tRNA
synthetases.
|
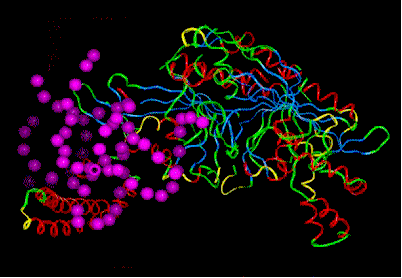
Picture to the right: Seryl-tRNA-Synthetase, in complex with tRNA. (only the phosphate residues - lilac balls - of the tRNA molecule are shown).
tRNAs have thus an adapter-function between mRNA and amino acids in protein synthesis. How many different tRNA molecules occur in a cell? The theoretical minimal value would be 20 corresponding to the number of different amino acids incorporated into proteins. The maximal value would be 61 (= 64 - 3), since no tRNA exist for the three stop codons. The actual number is roughly in the middle between these two theoretical assumptions. Different tRNAs have been proven to exist for a number of amino acids, but one tRNA is often able to recognize several codons as long as they differ only in the third base (wobble).
This leads to incorrect though thermodynamically stable pairings. Pairing is exact enough if the tRNA can distinguish between a purine and a pyrimidine in the third position (see also the codon table).
A further complication supervenes in plant cells: nucleus, chloroplasts and mitochondria contain genetic information and in each of these compartments occurs gene expression and replication mostly independent of those in the others. The processes of the different compartments are tuned by control mechanisms. Both mitochondria and chloroplasts have an own set of tRNAs the genetic information of which is encoded in the mitochondrial and chloroplast DNA, respectively.
Ribosomal RNA is a structural component of ribosomes. Both the small and the large subunit of the ribosome contain one and two (three), respectively, rRNA molecules of different size. The size is usually indicated by the S-value (the sedimentation constant). This group of RNA, too, occurs in different sets in the cytosol, chloroplasts and mitochondria.
The rRNA molecules of the ribosomes within the cytosol are considerably larger than those of the chloroplasts. The size of the latter is best compared to that of procaryotic rRNA. From a number of rRNA types of different origins the nucleotide sequences are known and they can be used for the study of the relationship between them. These data constitute one of the most reliable proofs for the endosymbiontic hypothesis. The fact that rRNA forms secondary structures much more complicated than that of tRNAs is very helpful. The secondary rRNA structures of chloroplasts and blue-green algae show a large degree of correspondence.
The genetic information for the instruction of the rRNA in chloroplasts is transcribed in one piece. A subsequent processing breaks the transcript down into 23 S, 5 S, 4,5 S and 16 S rRNA.
Comparable mechanisms can be found in the syntheses of other RNA species and also in different compartments. For more data on rRNA see rRNA-www-Server
Eucaryotic RNA is by far more subject to post-transcriptional processing than that of procaryotes. Two modifications have already been mentioned:
| Capping and polyadenylation of mRNA and | |
|
| the formation of rare bases in tRNA. |
In addition the primary transcripts are always longer than the final products. Some genes consist of several segments. The coding segments are called exons (expressed segments), the in-between ones are called introns. The primary transcription product contains both: exons separated by introns.
Introns have to be removed before translation. A number of specific ribonucleases exist and at least part of them has the function to transfer primary transcripts (hnRNA) into their functional state (mRNA, tRNA or rRNA). The predominant part of the hnRNA synthesized in the nucleus is never transported into the cytosol but is degraded directly after synthesis. No decisive explanation exists for this high turnover but it is assumed that a large quantity of ribonucleoside triphosphates has to be kept ready for use. But the large number would lead to quite an osmotic pressure. This could be decreased to physiological values by polymerization.
mRNA occurs in the cytoplasm never in a free state. It is always bound to specific proteins forming a ribonucleoprotein complex (RNP).
The DNA of the nucleus is usually very condensed. It is therefore not that easy to see the transcription units in an electron microscope. But a number of cells with lamp brush chromosomes has nuclei with less condensed DNA. A number of loops expands from a central chromosomal axis. Lamp brush chromosomes can be found in the oocytes of amphibians but also in the cells of the green alga Acetabularia mediterranea. On the free DNA transcription units can be detected. The following statements are based on the evaluation of electron microscopic images:
Transcription units may be of differing lengths but equal units occurring in tandem exist, too.
Transcription units can have opposite polarities. This is caused by the fact that in one case one strand is used and in the other the other strand. It is the position of the promoter that is decisive. The promoter is the sequence of nucleotides that is necessary for the binding of the DNA dependent RNA polymerase. It forms the starting point of transcription.
Between the transcription units are non-transcribed spacers of differing lengths.
The transcripts seem to be much shorter than the templates. This is caused by the secondary structure of the RNA (palindromes = hair pin structures) formed immediately after synthesis or/ and by complexes formed with proteins. Both mechanisms result in a drastically reduced length of the molecule.
Genes may have rates of transcription ranging between very high and rather low.
Polymerases are enzymes necessary for the formation of polynucleotides. Most of them are dependent on DNA as a template. Such RNA-polymerases bind to DNA regions called promoters. Transcription is finished as soon as the polymerase reaches a termination sequence. It is subsequently set free again. Repressors are proteins that bind tightly to the DNA (when active) and thus prevent transcription. RNA polymerases of pro- and eucaryotes differ fundamentally.
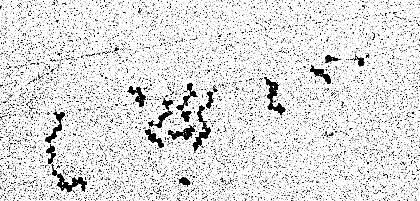
Transcription and translation of a DNA segment of Eschericia coli. The DNA looks like a thread. It is transcribed by several RNA polymerases simultaneously. In the picture the direction of transcription can be recognized by the lengths of the synthesized RNA (the lateral branchings). As soon as part of the RNA has been synthesized it is bound by ribosomes. The number of bound ribosomes grows with the length of the RNA. The ribosomes use the RNA as a template for protein synthesis (translation). (D. L. MILLER, Charlottesville, 1970).
Nuclear DNA is transcribed by three different classes of RNA-polymerases. All consist of several subunits. In many cases the molecular weight is known. All values hint at a considerable variability of the enzymes. They show that these proteins have changed rather significantly during evolution. The three classes (polymerase I, II and III) have hardly anything in common.
A further type of RNA polymerase exists in chloroplasts. It is related to the procaryotic RNA polymerase though it is encoded by a nuclear gene, and another RNA polymerase can be expected to be found in mitochondria. It has already been isolated from animal mitochondria.
Studies of Eschericia coli have shown that a transcription unit, a so-called operon, contains a number of signals besides the templates for the transcripts. Among them are a start- and a stop signal that mark beginning and end of the operon. Moreover exist binding sites for proteins that enhance, reduce or even inhibit the transcription of certain segments.
The signal effect is usually caused by an interaction of a protein (effector: activator or repressor) and a defined nucleotide sequence. Some effectors can switch between an active and an inactive state.
Many promoter regions of eucaryotic genes harbour a highly conserved AT-rich sequence at a distance of about 25 - 32 base pairs in front of the proper initiation point of the transcription, known as TATA-Box. It acts as a binding site for a transcriptional factor, the TATA-Box-binding protein. The specific binding of this protein to the DNA ensures a correct positioning of RNA-Polymerase II, that is required for the the transcrioption of protein-coding genes.
The change of an activity state can be induced by metabolites or nutriments (like, for example, a certain sugar). Comparable control mechanisms are also postulated for eucaryotes though they could not yet be proven to exist in detail (as they have for bacterial operons). Further details will be discussed when talking about differentiation, photomorphogenesis and hormone effects.
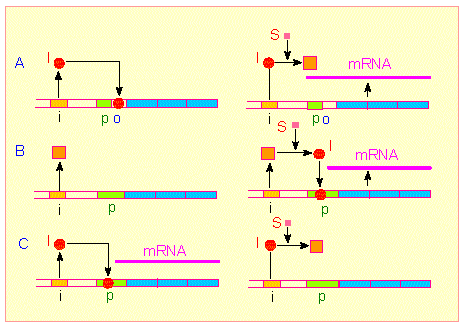
Three different control mechanisms of gene regulation. It is distinguished between regulator genes (ochre) and structural genes (light blue). A group of structural genes is preceded by a promoter (green: starting site of the RNA polymerase) and an operator (o: binding site of a regulator). Upper pictures (A): Substrate-induced control of transcription. The repressor encoded by the regulator gene binds to o and prevents the transcription of the structural genes. It can be inactivated by a substrate thus allowing transcription (lilac line: newly synthesized RNA). The enzymes encoded by the structural genes are involved in the breakdown of the substrate (JACOB-MONOD-model). Pictures in the middle (B): An inactive inducer is encoded by the regulator gene. The transcription rate of the structural genes is low. The inducer is activated by a substrate, binds to the promoter and enhances thus the activity of the RNA-polymerase: The rate of transcription is increased. Lower pictures (C): The regulator gene encodes an active inducer, the transcription rate of the structural genes is high. The inducer is deactivated by the binding of a substrate molecule.